Synthetic Cellular Engineering
Why Build Synthetic Cells from Scratch? The Bottom-Up Synthetic Biology Approach
How did life emerge from non-living matter on early Earth? Can we recreate this transition in the lab? These fundamental questions drive our research in bottom-up synthetic biology. By systematically reconstructing functional molecular entities, components, and modules, we aim to build synthetic cells that offer new insights into the mechanisms of life. The fact that life exists today on Earth demonstrates that it is possible to engineer life from matter. However, traditional top-down approaches, which rely on modifying existing biological systems, face inherent limitations, such as the complexity and unpredictability of natural cellular systems, as well as off-target effects that can compromise therapeutic outcomes. Therefore, engineering synthetic cells from the bottom up offers a more controlled and modular approach, allowing for the precise design and assembly of cellular components with tailored functions for applications in medicine, biotechnology, and beyond.
Key research interests
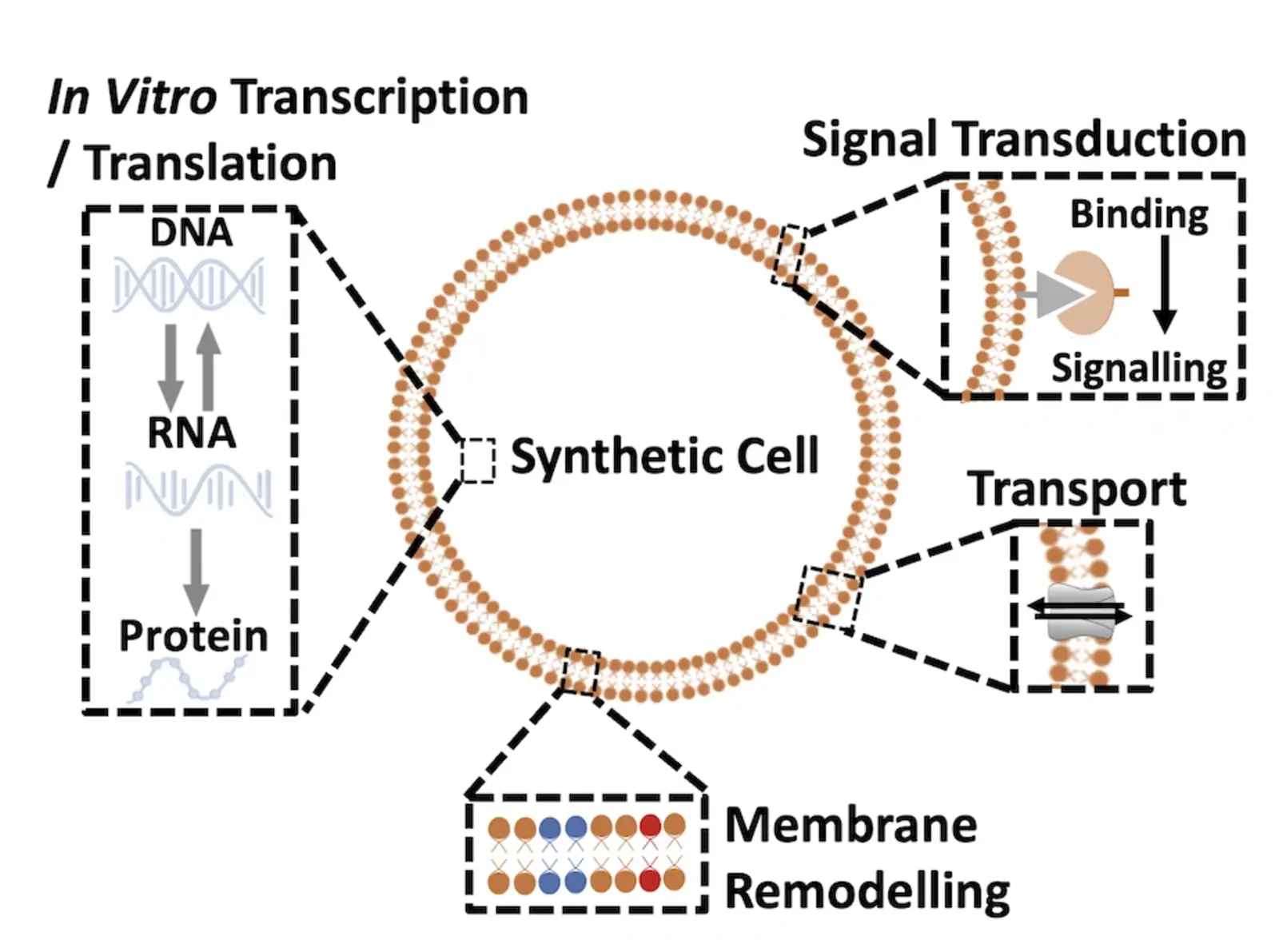
My lab focuses on designing and engineering synthetic cells with life-like functionalities using non-living molecular components. We integrate principles from synthetic biology, bioengineering, and nanotechnology to construct minimal cell models capable of complex biological processes. Key research areas include:
1) Signal Transduction: Engineering membrane receptors and pathways to enable controlled communication and response to external stimuli.
2) Molecular Transport: Developing selective transport mechanisms to regulate molecular cargo exchange across membranes.
3) Membrane Remodeling: Designing lipid bilayers with dynamic properties for shape transformations and functional modifications.
4) In Vitro Transcription & Translation: Reconstituting gene expression within synthetic cells to produce functional proteins.
Understanding Signal Transduction in Minimal Synthetic Cells
Signal transduction across cell membranes is fundamental to cellular function, influencing processes essential for health and disease. In immune cells, these pathways enable the recognition of pathogens and the activation of immune responses. However, dysregulation in signal transduction is a key factor in many diseases. Replicating these complex mechanisms in synthetic models presents both challenges and opportunities for advancing our understanding of cellular communication. My research group focuses on engineering synthetic immune receptors, along with transporter molecules, to replicate immune signaling with precise spatiotemporal control on the membranes of Giant Unilamellar Vesicles (GUVs). Light serves as a precise external trigger to manipulate synthetic cell functions with high spatiotemporal control, enabling dynamic regulation of gene expression, signaling pathways, and membrane remodeling with minimal invasiveness. By integrating optogenetics and nanotechnology, we aim to reconstruct and study immune cell membrane signal transduction in minimal synthetic cells. Through this bottom-up approach, our research seeks to uncover fundamental principles of cellular communication while paving the way for novel therapeutic strategies.
Expression and Translocation of Functional RNA and Proteins in Synthetic Cells
My other research focuses on developing a cell-free platform for the expression of immunogenic RNA aptamers, peptides, functional proteins, and immune receptors within synthetic cells built from non-living components. This approach provides a powerful tool for studying fundamental gene expression and regulation mechanisms, offering deeper insights into the molecular foundations of life. Beyond expression, we aim to investigate the translocation of larger biomolecules—such as RNA and proteins—across the phospholipid membranes of synthetic cells. A key challenge in synthetic cell engineering is enabling these molecules to move from the inner to the outer membrane, where they can interact with their environment, including targeted cancer cells. By designing mechanisms for controlled molecular translocation, we seek to facilitate direct interactions with cancer cell surfaces, triggering apoptosis via signal transduction pathways. This strategy presents significant advantages for studying protein-protein interactions. Unlike traditional phage display, which relies on viral membranes, our approach allows proteins to function in their native state within lipid bilayers, providing a more physiologically relevant system for understanding molecular recognition and therapeutic applications.
Find this exciting? We welcome applications from motivated, talented, and collaborative individuals at all levels with backgrounds in biology, biotechnology, biochemistry, chemistry, bioengineering, or related fields. Students with their own fellowships are especially encouraged to apply, and we would be happy to host them. Interested candidates can email (tchakrab@em.uni-frankfurt.de) their resume along with a motivation letter.

Contact:
Dr. Taniya Chakraborty
Independent Junior Group Leader
Institute of Biochemistry,
Biocenter N220 | room 1.04
Goethe-University Frankfurt
Max-von-Laue-Str. 9
D-60438 Frankfurt/M., Germany
Phone: +49-(0)-69 - 798 29477
Email: t.chakraborty(at)em.uni-frankfurt.de
ORCID: 0009-0005-4465-8343
X: @TaniyaChkrabort
