Membrane Biochemistry
In general, we focus on the molecular mechanisms of bacterial membrane transport systems and particularly on potassium-translocating systems. K+ homeostasis in bacteria is essential for the survival of an individual cell and of a bacterial community. Among others, K+ uptake and release are required for osmoadaptation upon host invasion and electrical signalling within a bacterial biofilm. Thus, several bacterial K+ translocation systems have been suggested as general pathogenicity factors and are potential targets for newly developed drugs. We are aiming at elucidating the proteins’ mechanisms of K+ translocation as well as identifying molecules that regulate their function. Currently, we are focusing on three major topics: The molecular mechanisms of bacterial osmoadaptation, the control of potassium homeostasis by the second messenger cyclic di-AMP, and the role of potassium channels in bacterial cell-cell communication.
Potassium transport by the chimeric pump KdpFABC
K+ uptake at very low extracellular concentrations is essential to the survival of microorganisms, and important to a wide variety of pathogens. Many bacteria and archaea rely on the KdpFABC complex to fulfill this task. KdpFABC is unique for combining an active P-type ATPase pump (KdpB) with a channel-like protein from the Superfamily of K+ Transporters (KdpA), alongside the auxiliary subunits KdpF and KdpC. Together, these subunits form a chimeric active uptake system that ensures cellular survival under K+ limitation.
Considering the subunits from two very different classes of membrane translocators, KdpFABC poses a number of interesting questions: How does K+ pass through the complex during transport? How exactly is translocation of K+ coupled to the ATP hydrolysis in KdpB, required for active transport? What is the role of each subunit in the complex? How does the regulation of KdpFABC work? We approach these questions with a combination of biochemical methods, structural work (cryo-EM), and MD simulations. In addition to illuminating a unique membrane transporter complex, answering these questions could help understand an important process in bacterial pathogenesis. (Contact: Sophie Ketter)
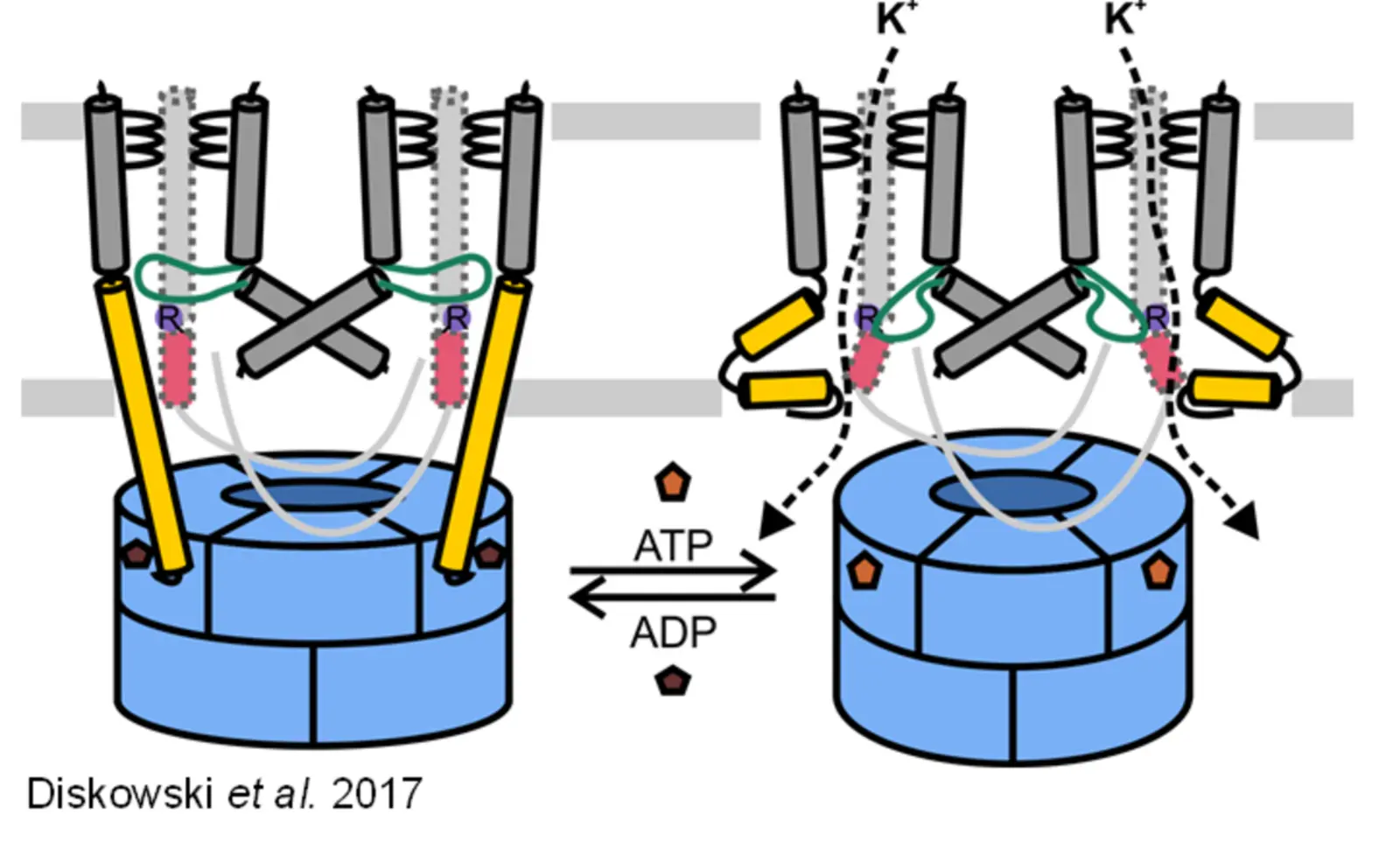
Molecular mechanism of potassium uptake through the channel KtrAB
Maintaining K+ homeostasis under osmotic stress is essential to the survival of microorganisms. The KtrAB channel, a complex of the dimeric channel subunit KtrB and the soluble cytosolic octameric ring of KtrA, is responsible for the K+ uptake under hyperosmotic stress. It has been shown that the opening and closing of the channel is triggered by the binding of ATP and ADP to the KtrA ring, respectively. It has been proposed that nucleotide binding would be followed by structural rearrangements in the KtrA ring and the breaking of an extended helix in KtrB, which previously reached into the cytoplasmic KtrA ring. As a result, the intramembrane loop, which is responsible for channel gating, gets more flexible and allows potassium flux. Nevertheless, the exact activation mechanism of KtrAB remains elusive which poses different questions: Is the nucleotide binding the only regulating trigger to open the channel? How are the structural rearrangements passed on from the KtrA ring to the KtrB dimer? Is there another trigger to close/inactivate the channel to prevent large uncontrolled potassium fluxes? We try to answer these questions using a mix of structural (cryo-EM) and biophysical (EPR spectroscopy) methods in addition to biochemical and functional assays as DSF, ITC, whole-cell potassium uptake assays and electrophysiology. (Contact: David Griwatz)
Characterization of compatible solute transporter EctI
The accumulation of compatible solutes is an important cellular adaptation mechanism to cope with osmotic stress, temperature stress, pH fluctuations, draught and UV-radiation. Compatible solutes, also known as chemical chaperones, comprise sugars, amino acids and amino acid derivatives as well as polyols and are uncharged and highly water-soluble compounds. Accumulation of compatible solutes can occur up to molar concentrations without interfering with cellular functions, hence the term “compatible”. They can be synthesized de novo or taken up via specific transport systems. EctI is such a transport system for the compatible solute ectoine. Ectoine is a very fascinating compatible solute, as it became medically relevant during the past years. Due to its characteristic to protect cells against UV-radiation, it is used in sunscreens. It also possesses anti-inflammatory properties, which is the reason for its usage in treatments of inflammatory skin and eye diseases. To this point, only a few transport systems for ectoine are identified and even less of them are thoroughly characterized. EctI is a secondary active transporter, belonging to the Sodium Solute Symporter (SSS)-family, which uses the inwardly directed electrochemical Na+ gradient to transport its actual substrate, ectoine, against the concentration gradient. As a member of the SSS-family, EctI adapts the LeuT-fold which comprises a 10-helix core with a 5+5 inverted repeat and two broken helices which play a mayor role in ion and substrate binding and translocation. For EctI it is unknown how the coupling of sodium ions to ectoine transport works, therefore I established in vitro assays to assess the kinetic mechanism of transport, to decipher the coupling stoichiometry and to identify the residues involved in substrate and ion translocation. Those assays are also used to identify possible additional substrates or inhibitors. In addition to the functional studies, structural analysis using X-ray crystallography and cryo-EM are performed to get further insights into the architecture of the protein. (Contact: Yvonne Hellmich)
Regulation of potassium uptake by the proton/potassium symporter KimA
Adaption to environmental changes is key for survival. Especially single cell organisms are exposed to drastic changes in their surrounding conditions. To keep the potassium homeostasis is critical to overcome osmotic fluctuations as well as maintain the whole cellular metabolism. KimA is part of the KUP (K+ Uptake) protein family. As a secondary active potassium transporter is KimA exploiting the proton motif force to take up potassium against its gradient even at low extracellular concentrations. KimA builds a homodimer in the plasma membrane whereby each monomer consists of a transmembrane part forming a pore and a cytoplasmic domain involved in regulation. The potassium uptake is inhibited by the cyclic nucleotide cyclic-di-AMP, which acts a newly discovered secondary messenger in bacteria. Still, it remains elusive how the regulation takes place on a molecular level. Which structural changes are triggered upon ligand binding? What are crucial amino acids relevant to fulfil inhibition? What cellular conditions are required for ligand binding? To mention only few open questions. We are trying to find answers by using structural biology (Cryo-EM/X-ray crystallography) as well as biochemical in vivo assays and in vitro assays (Ligand binding, protein purification etc.) approaches. (Contact: Michael Fuss)
Potassium-mediated Signaling in Biofilms
Bacterial biofilms can be found ubiquitously in a variety of environments from deep sea vents to pipes of the drinking and sewage water system or medical indwelling devices and open wounds. Especially in the latter, biofilms impose a great threat to humanity by contributing largely to the development of antibiotic resistance and virulence. Initial studies have revealed an involvement of the putative potassium channel YugO in the development and upkeep of biofilms formed by the gram-positive soil-dwelling bacterium Bacillus subtilis. By sending waves of potassium throughout the biofilm, the cells in the interior of the biofilm can communicate their need for nutrients and induce a periodic halting of growth of the peripheral cells which allows nutrients to diffuse deeper into the biofilm. This ensures the survival of the cells in the interior of the biofilm and increases overall viability. Recently, a similar signaling process was also observed for Escherichia coli involving the potassium channel Kch. However, while this phenomenon was investigated on a community level, open questions remain as to how this signaling process is achieved on a molecular basis. How is the opening of the potassium channels achieved? Which ligands control channel gating in response to the metabolic status of the cells? What is the role of the membrane potential in the signaling process and how do changes of intracellular potassium content and membrane potential influence biofilm viability? We are aiming at answering these questions using both, YugO from B. subtilis and Kch from E. coli, with a wide range of in vivo approaches including microfluidic studies of growing biofilms, the introduction of rhodopsins for targeted manipulation of the membrane potential and in vitro approaches using biochemical assays such as DSF, ITC and electrophysiology as well as structural studies including X-ray crystallography and cryo-EM. Unravelling the molecular mechanisms of the signaling process could open the opportunity to a new field of antibiotic targets and therapeutical applications in the long run. (Contact: Celina Thiel (YugO), Oliver Thon (Kch))
Contact
Prof. Dr. Inga Hänelt
Biocenter N200/1.08
phone +49-69-798 29 255
haenelt@biochem.uni-frankfurt.de