MHC I Peptide-loading and Chaperone Complexes at Onset of Adaptive Immunity
Identifying and eliminating infected or malignantly transformed cells are fundamental tasks of the adaptive immune system. For immune surveillance, the cell's metastable proteome is displayed as short peptides on MHC I molecules to cytotoxic T-lymphocytes. Interestingly, viruses have evolved immune evasion strategies, which target the antigen presentation pathway. Our knowledge about the track from the proteome to presentation of peptides has greatly expanded in recent years and has lead to a comprehensive understanding of antigen presentation and how viruses escape immune surveillance.
As a highly dynamic supramolecular complex in the ER membrane, the peptide-loading complex (PLC) orchestrates peptide translocation, loading, and editing of MHC I molecules. The PLC assembles around the heterodimeric ATP-binding cassette (ABC) transporter TAP, two copies of tapasin, ERp57, calreticulin, and MHC I/β2m heterodimers. The disulfide-linked complex between tapasin-ERp57 forms the minimal functional unit of the PLC, which facilitates peptide loading onto MHC I and editing of the peptide repertoire. Strikingly, classical and nonclassical MHC I allomorphs are associated with the PLC, suggesting that most allomorphs traverse through the PLC on their way to the cell surface.
We have determined the structure of native PLC isolated from Burkitt's lymphoma cells using an engineered viral inhibitor. Structures of PLC at distinct assembly states provide insights into MHC I quality control and unveil the molecular details underlying the onset of an adaptive immune response. In the long term, we are addressing fundamental questions of adaptive immunity to advance our knowledge on the sophisticated cellular proofreading machinery and to elucidate mechanisms of viral immune evasion by an interdisciplinary approach including biophysics, biochemistry, structural biology and genome engineering strategies.
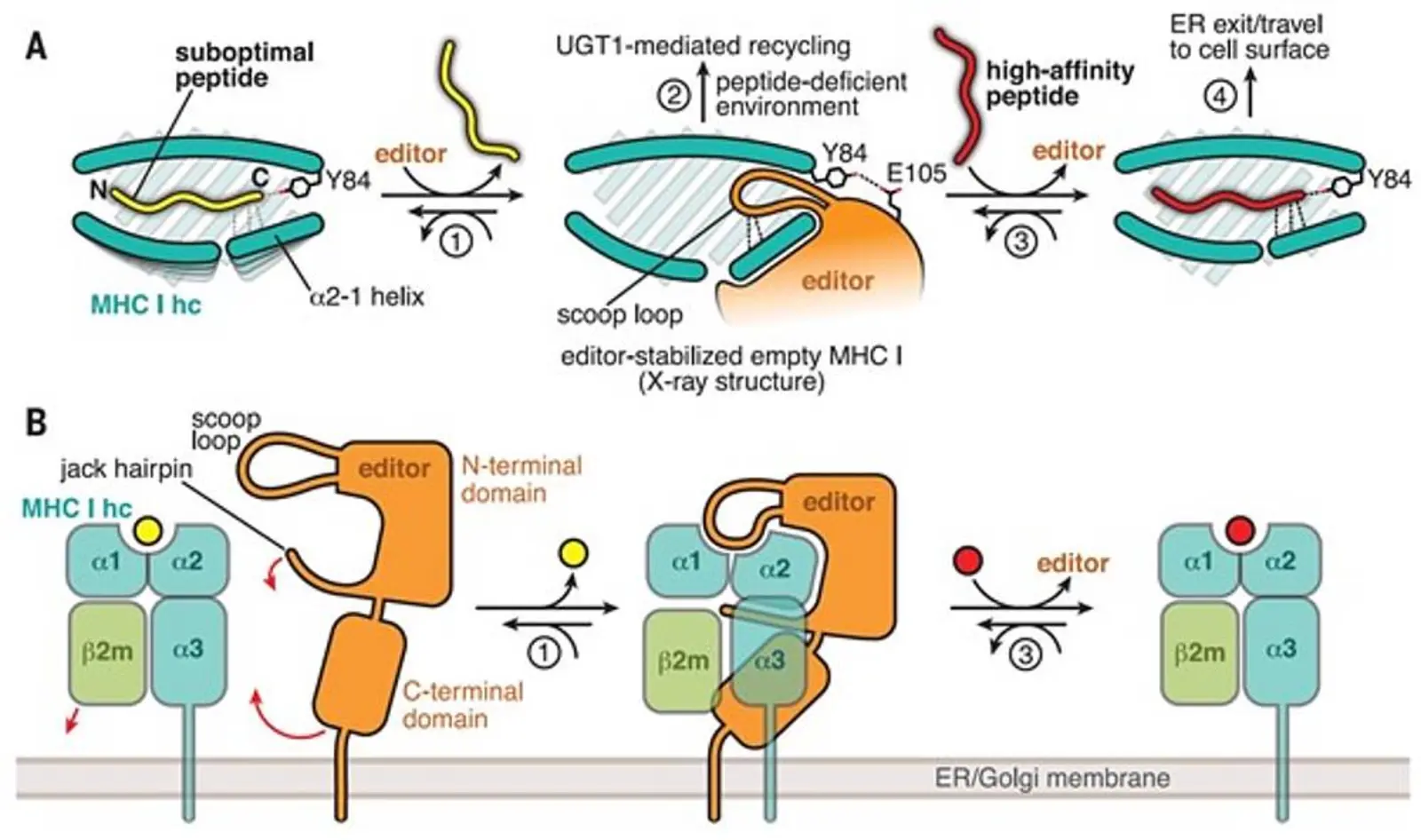
Specialized cells of the immune system can detect viral infection and malignant transformation by recognizing aberrant peptides presented on major histocompatibility complex class I (MHC I) molecules at the surface of diseased cells. The displayed peptides are representative fragments of the cellular proteome and are shuttled into the endoplasmic reticulum (ER) where the loading onto MHC I takes place. This loading is catalyzed by the two MHC I-specific chaperones tapasin and TAP-binding protein-related (TAPBPR). During loading, the peptides are selected for their ability to form stable peptide-MHC I (pMHC I) complexes, a process called peptide editing or proofreading. The stability of the pMHC I complexes is of pivotal importance in eliciting an effective immune response.
We have been able to elucidate the mechanistic basis of chaperoning and peptide proofreading by determining the crystal structure of TAPBPR in complex with MHC I. The structure shows that TAPBPR stabilizes the malleable MHC I in a peptide-receptive conformation by forming an extensive interface that includes and perturbs structural elements crucially involved in peptide binding. The peptide-binding groove of MHC I adopts a widened conformation in the complex which facilitates peptide exchange and selection of high-affinity ligands.
We are currently using biochemical, biophysical and structural approaches to further unravel the intricacies of peptide editing. A comprehensive understanding of this central process in immunity is expected to facilitate vaccine development in the fight against infections and tumors.
Publications:
Sagert L, Hennig F, Thomas C, Tampé R
A loop structure allows TAPBPR to exert its dual function as MHC I chaperone and peptide editor.
2020, eLife, 9, e55326; doi: 10.7554/eLife.55326
Trowitzsch S, Tampé R
Multifunctional chaperone and quality control complexes in adaptive immunity.
2020, Annu Rev Biophys, 49,135-161; doi: 10.1146/annurev-biophys-121219-081643
Thomas C, Tampé R
MHC I chaperone complexes shaping immunity.
2019, Curr Opin Immunol, 58,9-15; doi: 10.1016/j.coi.2019.01.001
Blees A, Januliene D, Hofmann T, Koller N, Schmidt C, Trowitzsch S, Moeller A, Tampé R.
Structure of the human MHC-I peptide-loading complex.
Nature. 2017 Nov 23;551(7681):525-528. doi: 10.1038/nature24627. Epub 2017 Nov 6.
- Cover Story and News&View in Nature 551,442-3
- Dispatch in Curr Biol 28,R83-R86
- Highlight in Faculty 1000 Prime
Thomas C, Tampé R.
Structure of the TAPBPR-MHC I complex defines the mechanism of peptide loading and editing.
Science. 2017 Nov 24;358(6366):1060-1064. doi: 10.1126/science.aao6001. Epub 2017 Oct 12.
- Insights&Perspectives in Science 358,992-3
- News&View in Nature 551,442-3
- Viewpoint in Biochemistry 57,1423-5
- Highlight in Faculty 1000 Prime
Thomas C, Tampé R
Proofreading of Peptide-MHC complexes through dynamic multivalent interactions.
2017, Front Immunol, 8,65; doi: 10.3389/fimmu.2017.00065
Fleischmann G, Fisette O, Thomas C, Wieneke R, Tumulka F, Schneeweiss C, Springer S, Schäfer LV, Tampé R
Mechanistic basis for epitope proofreading in the peptide-loading complex.
2015, J Immunol, 195,4503-13; doi: 10.4049/jimmunol.1501515
- Highlighted as Cutting Edge Article in J Immunol
PROJECT MEMBERS
Dr. Simon Trowitzsch
Group Leader
phone +49-(0)69-798-29274
trowitzsch@biochem.uni-frankfurt.de
Dr. Lukas Susac
Postdoctoral Fellow
phone: +49-(0)-69-798-29274
susac@em.uni-frankfurt.de
Darja Cernova
PhD Student
phone: +49-(0)-69-798-29502
cernova@em.uni-frankfurt.de
Amin Fahim
PhD Student
phone: +49-(0)-69-798-29502
fahim@em.uni-frankfurt.de