

Ribosome recycling in mRNA translation and protein quality control
Cellular protein biosynthesis is conserved and essential for life. Moreover, mRNA translation constitutes a key process at the crossover of numerous pathways and signaling cascades with the ribosome as a central regulatory hub. We investigate the role of ribosome recycling by ATP-binding cassette (ABC) protein ABCE1 as a crucial control step for the biogenesis of functional proteins and the degradation of aberrant nascent chains in protein quality control. These two crucial adjusting wheels define protein homeostasis and are directly linked to neurodegeneration and the adaptive immune response to viral infection and malignantly transformed cells.
ABCE1 is one of the most conserved proteins in evolution, ubiquitously found and essential in Archaea and Eukarya. After canonical translation termination, in mRNA surveillance and ribosome-associated quality control, as well as in ribosome biogenesis, assembled ribosomal complexes are recycled into their subunits by ABCE1 via an ATP-dependent power stroke. In our RNA team, we focus on the ATPase cycle of ABCE1 during ribosome recycling and the post-splitting complex as a platform for translation initiation.
Our integrated scientific approach comprises biochemical, biophysical and cell biological methods as well as structural biology. The methodical portfolio includes classical biochemistry, structural studies by cryogenic electron microscopy (cryo-EM) reconstruction and X-ray crystallography, single-molecule Förster resonance energy transfer (smFRET), chemical cross-linking and mass spectrometry (XL-MS) and in vivo mutational studies. We collaborate with high-profile international research groups to conduct interdisciplinary scientific projects and gain exciting insights into ribosome recycling, mRNA translation and protein quality control.
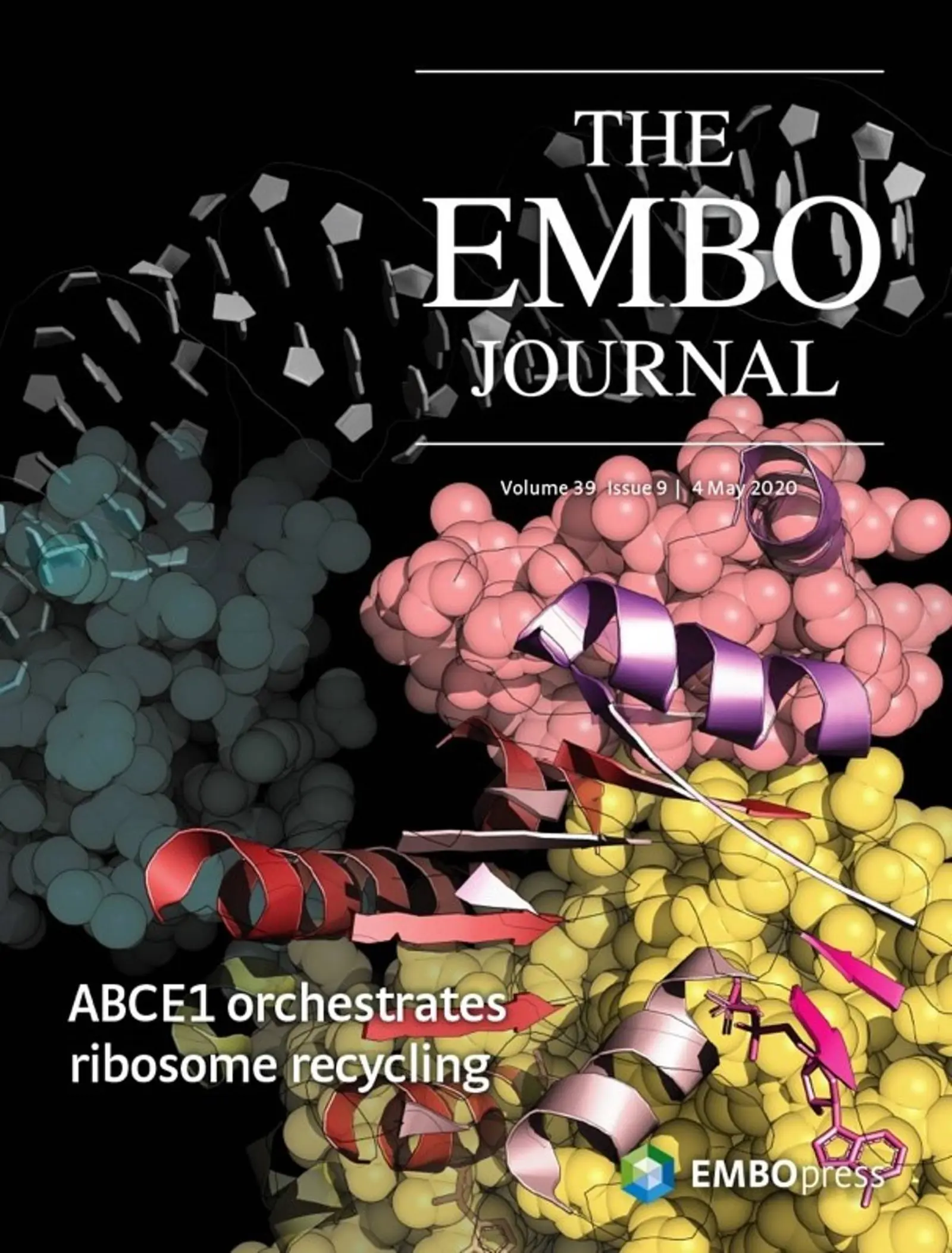
Ribosome recycling involves allosteric communication between ABCE1 and the ribosome
Our recent publication promoted on the front cover in May 2020 issue of the EMBO Journal constitutes the first high-resolution structure of the archaeal 30S ribosome and describes the molecular architecture of the ribosomal post-splitting complex and the allosteric crosstalk between the 30S subunit and ABCE1. Here, we continued our successful collaborations with the Beckmann Lab at Munich's LMU and Frankfurt's Institute of Molecular Bioscience to provide structural, biochemical and cell biological insights into ribosome recycling.
Check it out on Twitter and read the full story in the EMBO Journal.
Click here to view the work presented by Holger Heinemann at the 25th Annual Meeting of the RNA Society in May 2020.
Inner mechanistics of ABC proteins are defined by dynamic equilibria
In our smFRET study published in Cell Reports, we propose dynamic equilibria instead of static single-state conformational models to describe the mechanism of ABC-type ATPases. In collaboration with the Cordes Lab (LMU Munich) and the Gouridis Lab (KU Leuven), we labelled ABCE1 and recorded hundreds of ribosomal 30S and 70S complexes in single-molecule alternating laser emission and excitation (smALEX) setup to show that ABCE1 is plastic protein and the functional asymmetry of its two ATPase sites is defined by conformational equilibria.
Read the full story in Cell Reports.
Same same but different - two structurally similar ATP sites have distinct biological functions
In our publication featured on the front cover of Life Science Alliance, we dissected the complex process of ribosome recycling into discrete steps and finally resolved the enigma about the asymmetry in the two ATPase sites of ABCE1. We show that one site is a control site that serves as a ribosome recognition checkpoint and activates the other site, which is responsible for energizing ribosome splitting. Follow the links below to find out more about the hardcore protein and ribosome biochemistry routinely carried out in our lab to decipher biological processes, and to prepare and assess samples for biophysical and structural studies.
Check it out on Twitter and read the full story in Life Science Alliance.
Ribosome recycling and mRNA translation is an exciting, constantly growing research field
If you are interested in the big picture around the regulatory role of ribosome recycling, please enjoy our recent review published in the 401st Jubilee Issue of Biological Chemistry. Find out more about versatile ABC machines in RNA remodeling in our latest TiBS Review.
Publications:
Nürenberg-Goloub E, Kratzat H, Heinemann H, Heuer A, Kötter P, Berninghausen O, Becker T, Tampé R, Beckmann R
Molecular analysis of the ribosome recycling factor ABCE1 bound to the 30S post-splitting complex.
2020; EMBO J 39,e103788; doi:10.15252/embj.2019103788
Nürenberg-Goloub E, Tampé R
Ribosome recycling in mRNA translation, quality control, and homeostasis.
2019; Biol Chem, 401,47-61; doi:10.1515/hsz-2019-0279
Gouridis G, Hetzert B, Kiosze-Becker K, de Boer M, Heinemann H, Nürenberg-Goloub E, Cordes T, Tampé R
ABCE1 controls ribosome recycling by an asymmetric dynamic conformation equilibrium.
2019; Cell Rep 28, 723-634; doi:10.1016/j.celrep.2019.06.052
Gerovac M, Tampé R
Control of mRNA Translation by Versatile ATP-Driven Machines.
Trends Biochem Sci. 2019 Feb;44(2):167-180. doi: 10.1016/j.tibs.2018.11.003. Epub 2018 Dec 7.
Nürenberg-Goloub E, Heinemann H, Gerovac M, Tampé R
Ribosome recycling is coordinated by processive events in two asymmetric ATP sites of ABCE1.
Life Sci Alliance. 2018 Jun 14;1(3). pii: e201800095. doi: 10.26508/lsa.201800095.
-Highlighted as Front Cover Page
Heuer A, Gerovac M, Schmidt C, Trowitzsch S, Preis A, Kötter P, Berninghausen O, Becker T, Beckmann R, Tampé R
Structure of the 40S-ABCE1 post-splitting complex in ribosome recycling and translation initiation.
Nat Struct Mol Biol. 2017 May;24(5):453-460. doi: 10.1038/nsmb.3396. Epub 2017 Apr 3.
- Previews in Mol Cell 66, 578-80
Kiosze-Becker K, Ori A, Gerovac M, Heuer A, Nürenberg-Goloub E, Jan Rashid U,
Becker T, Beckmann R, Beck M, Tampé R
Structure of the ribosome post-recycling complex probed by chemical cross-linking and mass spectrometry.
Nat Commun. 2016 Nov 8;7:13248. doi: 10.1038/ncomms13248.
Project Members
Dr. Lukas Susac
Postdoctoral Fellow
phone: +49-(0)-69-798-29274
susac@em.uni-frankfurt.de
Dr. Simon Trowitzsch
Postdoctoral Fellow
phone +49-(0)69-798-29273
trowitzsch@biochem.uni-frankfurt.de
Tamina Sens
PhD Student
phone: +49-(0)-69-798-29471
sens@em.uni-frankfurt.de
Jonas Göhmann
PhD Student
phone: +49-(0)-69-798-29471
goehmann@em.uni-frankfurt.de
Selma Guenov
PhD Student
phone: +49-(0)-69-798-29471
s.guenov@em.uni-frankfurt.de
